Rat Cerebellum volume¶
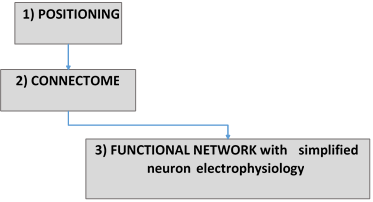
This Use Case is used to connect cerebellar neurons and can be found in Online Use Cases/Circuit Building/Connectome/Rat cerebellum volume.
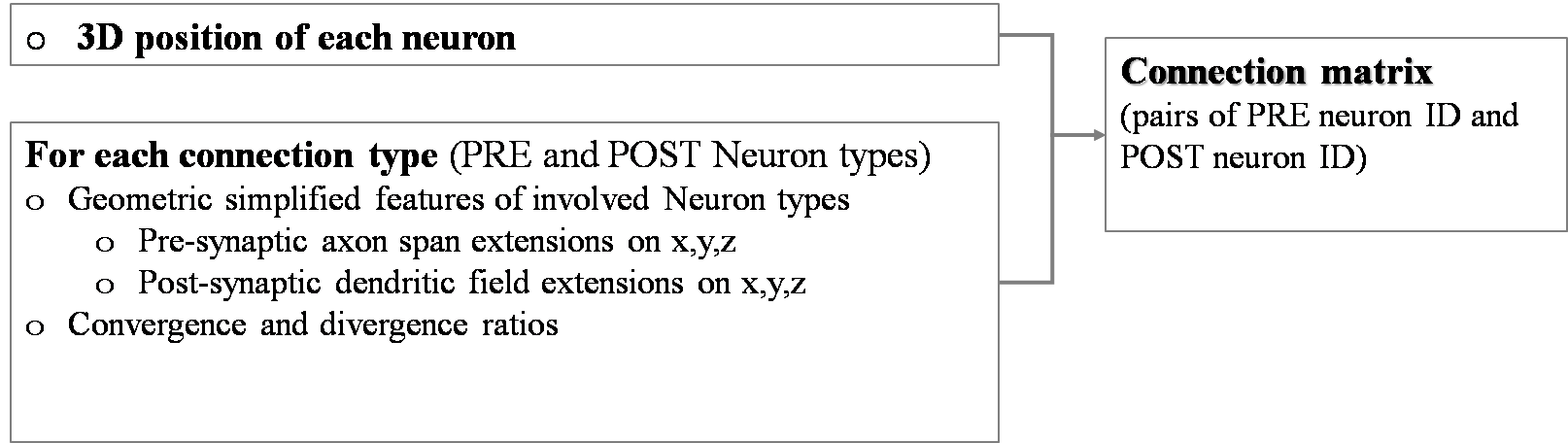
Approach: the potential connections are created by imposing geometric constraints (based on the anisotropic simplified geometric features of the pre- and post-synaptic neuron types). Then, if needed, a selection (pruning) is carried out by defining convergence or/and divergence ratios and applying them by distance-dependent probability.
Inputs:
hdf5 matrix (saved in /storage) obtained from the “Cell positioning” Use Case (found in Online Use Cases/Circuit Building/Cells placement/Rat cerebellum volume/).
The parameters are automatically imported from user-defined parameters filled in the GUI of the “Cell positioning” Use Case and from scaffold_params.py (in /storage).
Expert users can modify connection-specific parameters could be modified from scaffold_params.py (in /storage)
Convergence and divergence ratio of each connection type
Output:
Within the hdf5 file generated from the “Cell positioning” Use Case, a new key is added named “/connections”. It contains multiple items, one for each connection type (e.g. “/glom_grc”, “/glom_goc”,…). Each of them is made up of a matrix with 2 columns (PRE Neuron ID and POST Neuron ID). The enriched hdf5 file is saved in /storage.
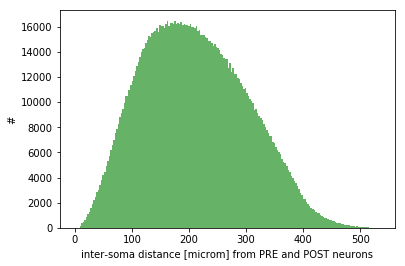
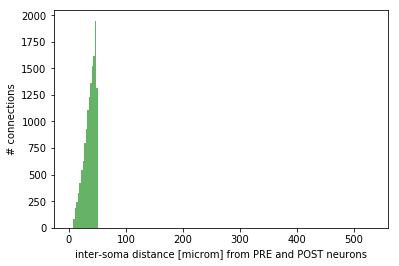
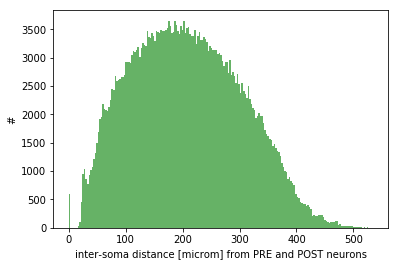
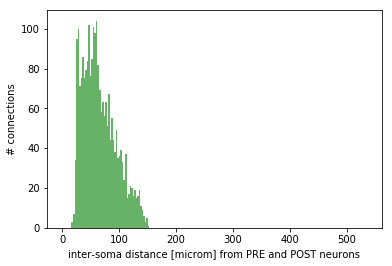
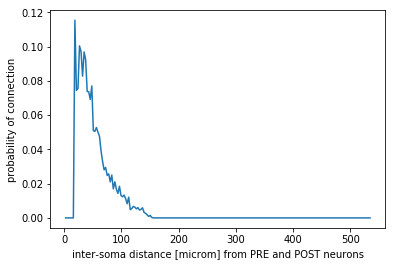
Monitoring: effective convergence and divergence ratios and pairwise distances among connected neurons (metrics_connectome.py in /storage) and probability of connection dependent on inter-soma distance between connected neurons (monitoring_probConn.py in /storage)
As an example, a 3D basic visualization of the connection from Glomeruli to Golgi cells is displayed. One PRE Neuron ID is selected, and all the POST neurons receiving its outward connections are plotted (Divergence). Also, one specific POST Neuron ID is selected, and all the PRE neurons sending connections to it are plotted (Convergence).
Additional information:
This whole Use Case should take about 4 minutes for a volume base of 400 x 400 µm. If enabled, the computation of the metrics for each connection type adds about 15 minutes for such a network size.
No login to any other computer required
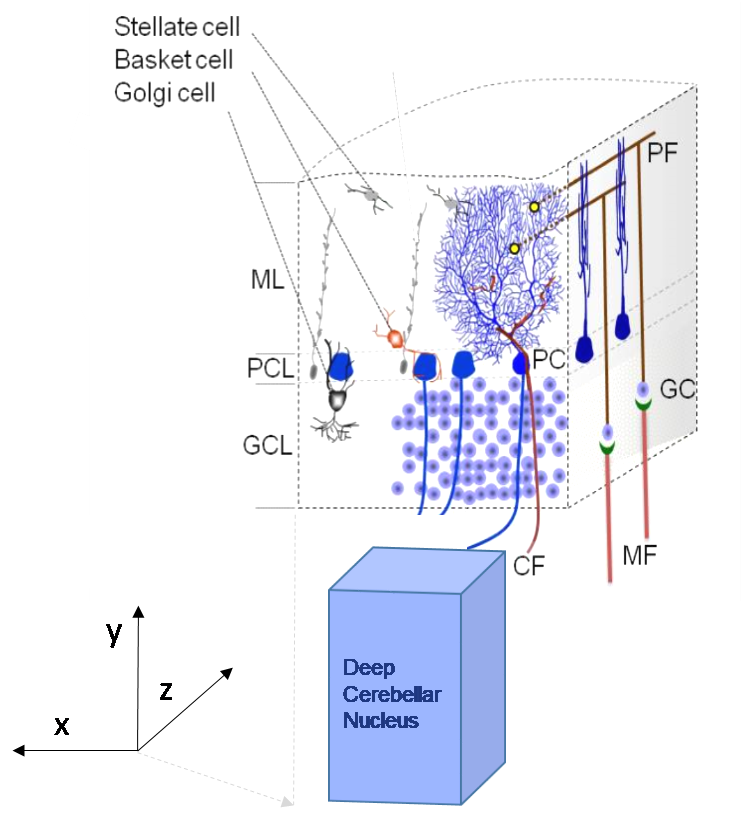
1.1) Glomeruli – Granule Cells (EXC)
- Requirements imposed in the code:
Simple connectivity rule: a single GrC cannot send more than one dendrite into the same glomerulus
Convergence: 4
The maximal length of GrC dendrites is ~ 40 µm (average = 13.6)
- Target values to be matched:
Divergence: ~53
Mean number of dendrites per Granule Cell: 4 (+/- 1)
1.2) Glomeruli – Golgi Cells (basolateral dendrites) (EXC)
- Requirement imposed in the code:
Extent of GoC basolater arborization (under the soma) is ~ 100 µm (diameter)
- Target values to be matched:
Divergence: 4
Convergence: 40 (on GoC basal dendrites) (not the same Glomeruli on the same GoC!)
Number of GoC (basal) dendrites = 4.1 +/- 1.4
1.3) Golgi Cells – Glomeruli (INHIB)
- Requirements imposed in the code:
Divergence: 40
Convergence: 1 (max)
Anisotropic extension of GoC axonal plexus: ~150 µm on x axis; ~150 µm on y axis; ~30 µm on z axis
- Target values to be matched:
The GoC axon inhibits granule cells dendrites inside the glomeruli.
1.4) Granule Cells by Ascending Axons - Golgi Cells (EXC)
- Requirements imposed in the code:
Divergence: 1 (max)
Convergence: 400
Extent of GoC arborization (sphere) is ~ 100 µm (diameter)
1.5) Granule Cells by Parallel Fibers - Golgi Cells (apical dendrites) (EXC)
- Requirements imposed in the code:
Convergence: 1600, of which 400 are from the nearest granules already connected by Ascending Axons from 1.4) connection type
GoC apical dendritic tree is ~ 100 µm (diameter)
- Target value to be matched:
Divergence: ~2
2.1) Stellate Cells – Purkinje Cells (INHIB)
- Requirements imposed in the code:
Anisotropic extension of SC axon: ~500 µm on x axis; ~100 µm on z axis
Divergence: 2
Convergence: 20
Activated BEAM ON (even the PCs excited by the Ascending Axons bundle are within the potential inhibition from the Molecular Layer interneurons, i.e. Stellate and Basket Cells)
2.2) Basket Cells - Purkinje Cells (INHIB)
- Requirements imposed in the code:
Anisotropic extension of SC axon: ~500 µm on x axis; ~100 µm on z axis
Divergence: 2
Convergence: 20
Activated BEAM ON (even the PCs excited by the Ascending Axons bundle are within the potential inhibition from the Molecular Layer interneurons, i.e. Stellate and Basket Cells)
2.3) Stellate Cells - Stellate Cells & Basket Cells - Basket Cells (INHIB)
- Requirements imposed in the code:
Divergence: 4
Maximum inter-soma distance for connection (150 µm on x-y plane; 50 µm along z)
- Target value to be matched:
Convergence: 4
3.1) Granule Cells by Ascending Axons – Purkinje Cells (EXC)
- Requirements imposed in the code:
Divergence: 1
Anisotropic extension of PC dendritic tree (symmetrically around the soma): ~130 µm on x axis; ~3.5 µm on z axis; all molecular layer thickness (y-axis)
- Target value to be matched:
Convergence: 200-400
3.2) Granule Cells by Parallel Fibers – Purkinje Cells (EXC)
- Requirement imposed in the code:
Anisotropic extension of PC dendritic tree (symmetrically around the soma): ~130 µm on x axis
- Target value to be matched:
Divergence: 30 (but experimentally difficult on slices, since long PFs but cut)…
3.3) Granule Cells by Parallel Fibers – Basket Cells (EXC)
- Target values to be matched:
Basket cell dendritic “circle” (2D on x-y sagittal plane) of 15 µm radius
PFs into the Molecular Layer; from GrC soma, y shift of AA length (taken from this distribution 151 ± 66 µm, with a minimum depending on the GrC soma y-coordinate, in order to always reach the ML)
3.4) Granule Cells by Parallel Fibers – Stellate Cells (EXC)
- Target values to be matched:
Stellate cell dendritic “circle” (2D on x-y sagittal plane) of 15 µm radius
PFs into the Molecular Layer; from GrC soma, y shift of AA length (taken from this distribution 151 ± 66 µm, with a minimum depending on the GrC soma y-coordinate, in order to always reach the ML)
4.1) Purkinje Cells – Deep Cerebellar projection Neurons (INHIB)
- Requirements imposed in the code:
DCpN dendritic field (planar with random angle, extensions over the network volume)
Divergence: 4 or 5 (randomly)
- Target value to be matched:
Convergence: 34-52
EXAMPLE
From “Cells Placement” in the volume:
x = 400 µm, z = 400 µm (→ DCN 200 x 200 µm)
y = 930 µm (600+ 150+30+150 µm), i.e. thickness DCN + GRL+ PCL + ML
1.1) Glomeruli – Granule Cells (EXC)
Div = 50 ± 24
Conv = 4 ± 0
3D distance = 12 ± 5 µm
1.2) Glomeruli – Golgi Cells (basolateral dendrites) (EXC)
Div = 2 ± 1
Conv = 65 ± 28
3D distance = 37 ± 10 µm
1.3) Golgi Cells – Glomeruli (INHIB)
Div = 31 ± 14
Conv = 1 ± 0
3D distance = 37 ± 17 µm
2D distance sagittal (x-y) = 35 ± 18 µm
Distance on transverse direction (z) = 7 ± 4 µm
1.4) Granule Cells by Ascending Axons - Golgi Cells (EXC)
Div = 1 ± 0
Conv = 383 ± 58
3D distance = 30 ± 16 µm
1.5) Granule Cells by Parallel Fibers - Golgi Cells (apical dendrites) (EXC)
Div = 4 ± 2
Conv = 1600 ± 0
3D distance =148 ± 103 µm
2.1) Stellate Cells – Purkinje Cells (INHIB)
Div = 2.6 ± 1.6
Conv = 20 ± 0.1
3D distance = 227 ± 85 µm
2.2) Basket Cells - Purkinje Cells (INHIB)
Div = 2.6 ± 1.6
Conv = 20 ± 0.1
3D distance = 186 ± 94 µm
2.3) Stellate Cells - Stellate Cells & Basket Cells - Basket Cells (INHIB)
Div = 4 ± 0
Conv = 4 ± 2
3D distance = 66 ± 29 µm
3.1) Granule Cells by Ascending Axons – Purkinje Cells (EXC)
Div = 0 or 1 (20%)
Conv = 252 ± 11
3D distance = 98 ± 40 µm
2D distance sagittal (x-y) = 98 ± 40 µm
Distance on transverse direction (z) = 0.8 ± 0.5 µm
3.2) Granule Cells by Parallel Fibers – Purkinje Cells (EXC)
Div = 23 ± 3
Conv = 28827 ± 165
3D distance = 178 ± 79 µm
3.3) Granule Cells by Parallel Fibers – Basket Cells (EXC)
Div = 10 ± 6
Conv = 1425 ± 399
3D distance = 224 ± 65 µm
2D distance sagittal (x-y) = 224 ± 65 µm
Distance on transverse direction (z) = 6 ± 4 µm
3.4) Granule Cells by Parallel Fibers – Stellate Cells (EXC)
Div = 5 ± 6
Conv = 635 ± 436
3D distance = 222 ± 65 µm
2D distance sagittal (x-y) = 222 ± 65 µm
Distance on transverse direction (z) = 6 ± 4 µm
4.1) Purkinje Cells – Deep Cerebellar projection Neurons (INHIB)
Div = 4 or 5
Conv = 27 ± 3
3D distance = 535 ± 145 µm