Compare a specific region of the electrostatic potentials surrounding a set of protein isoforms with multipipsa¶
Overview¶
This use case describes how to compare the electrostatic potentials of different isoforms of a protein at a specific site on its surface and to cluster these isoforms by similarity of their electrostatic potentials in this region. It uses the multipipsa software tool, which helps to automate these calculations by providing a python wrapper for the following open source software tools:
PDB2PQR: A tool that takes a protein structure in PDB format, adds missing hydrogen atoms, and creates a structure file in PQR format. The PQR file format is derived from the PDB format for describing atomic data, but with the occupancy and temperature factor fields replaced with atomic partial charges and radii.
APBS: A tool that calculates electrostatic potentials through solution of the Poisson-Boltzmann equation, one of the most common continuum models for describing electrostatic interactions between molecular solutes in salty, aqueous media.
Background¶
For details on computation of the electrostatic potential see Calculate the electrostatic potential of a protein from its atomic structure.
For details on the similarity computations and multipipsa algorithm, see Compare the electrostatic potentials surrounding a set of protein isoforms with multipipsa
Input Data¶
In this use case, we use as our input structure a structure of the catalytic domain of the enzyme adenylyl cyclase 5 (AC5), modelled during the work described in Tong et al (2016). The structures of the AC isoforms were created via homology modelling using the same template. The region where there are significant structural differences between the isoforms is in a flexible loop region that was not defined in the template structure. There are also variations in sequence length across AC isoforms in this region.
Procedure¶
Structure of AC5 is visualized. The catalytic domain of AC5 is a dimer consisting of two protein chains. In the full structure of AC5 these two chains are connected by a series of transmembrane helices that anchor the protein in the post-synaptic membrane.
Then the Pdb2Pqr method is used to generate hydrogen atoms in the protein structure. Proteins contain a number of ionisable amino acids, which can exist in different protonation states, depending on the pH of the solution they are in. PDB2PQR can predict the states of these amino acids, at a given pH (defined as 7.4 in the last cell, a normal physiological pH), then add all missing hydrogen atoms to the structure, and assign atomic charges and radii to all atoms. By default, multipipsa assigns charges and radii from the Amber force field.
The APBS method used to solve the linearised Poisson-Boltzmann equation to obtain the electrostatic potential in the dx and UHBD file formats. It also creates a dx file describing the solvent excluded volume of AC5. This is used for visualisation later.
Finally, electrostatic potentials of isoforms are compared. In this use case, we want to limit our comparison the electrostatic potentials to a specific region. We do this by defining a residue within one of the AC isoforms. Here we choose AC5 as the reference and define the analysis region as a sphere of radius 10 Angstrom centered on the Cα atom of residue 32 within this reference structure.
Results¶
Structure of the AC5 dimer:
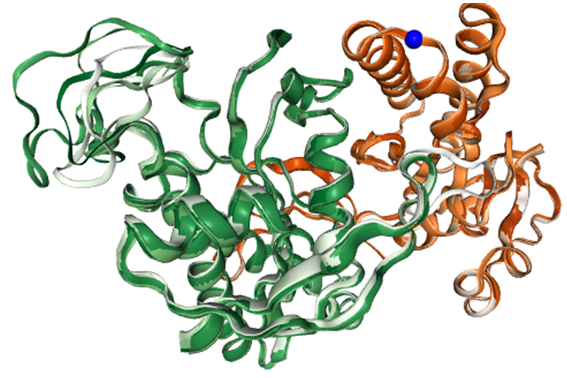
a blue sphere shows where the analysis is centered (the analysis region is defined as a sphere of radius 10 Angstrom). It lies on the edge of a groove formed by three helices (located at Cα atom of residue 32).
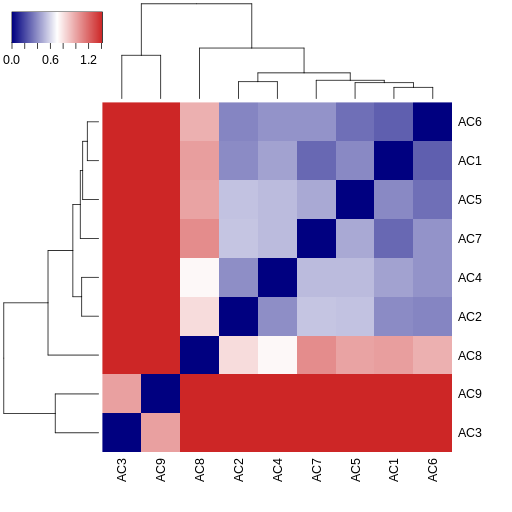
The MultiPIPSA analysis creates as output an image file showing the pairwise distances between AC isoforms as a 2D heatmap. The results are also clustered using a single linkage hierarchical method. The resulting dendograms are shown along the edges of the heatmap.